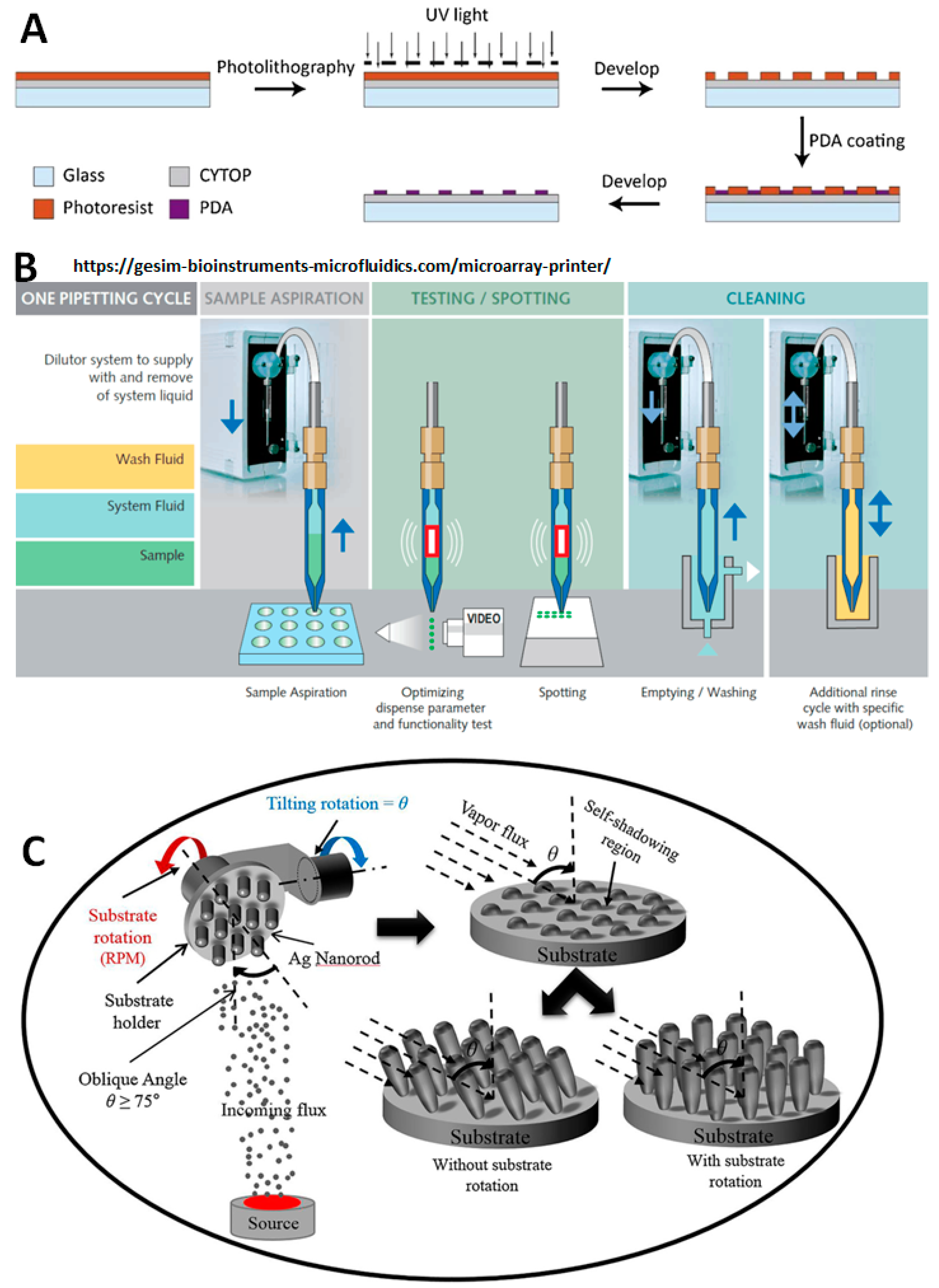
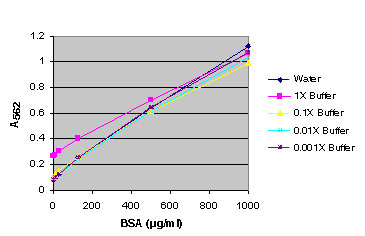
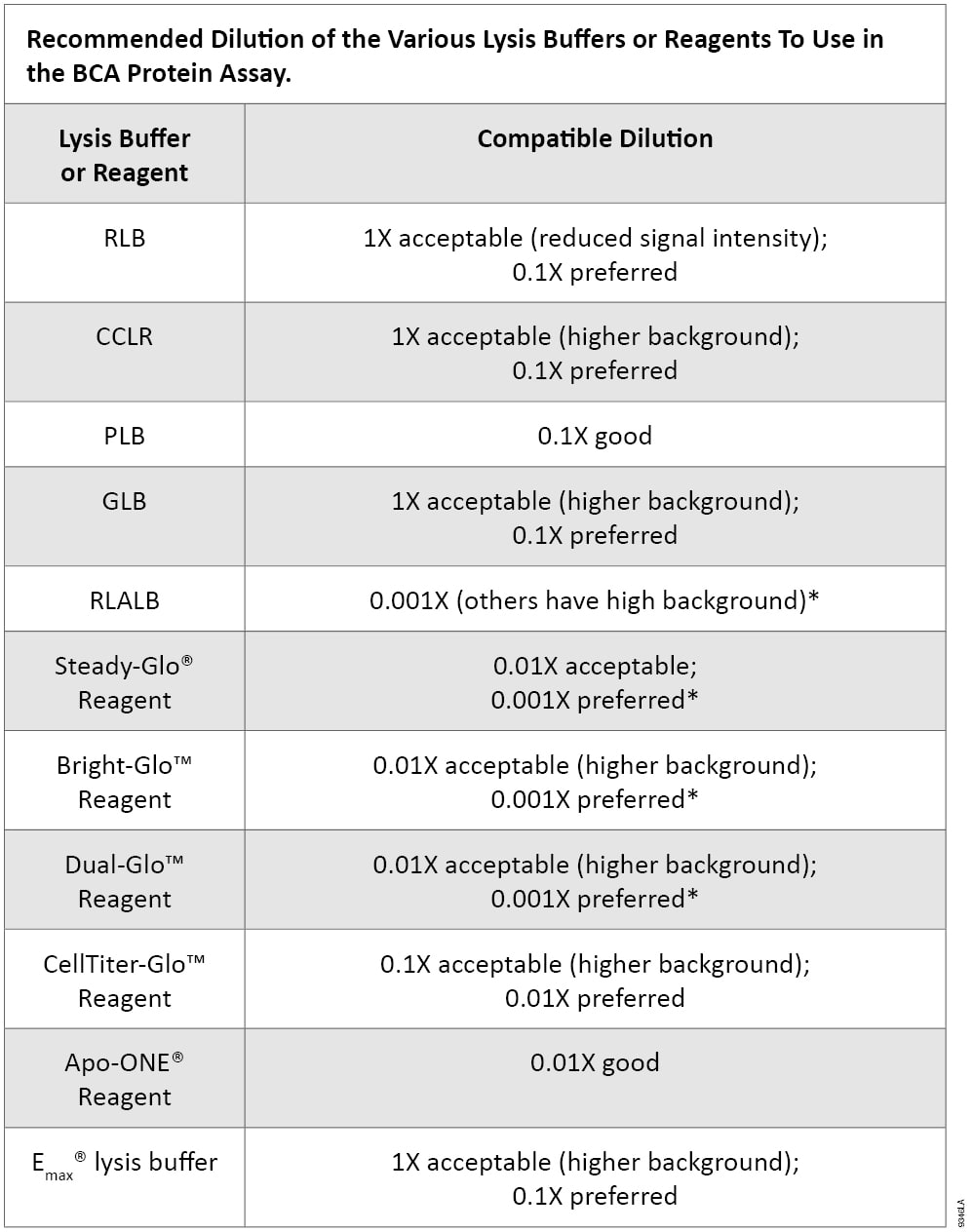
TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

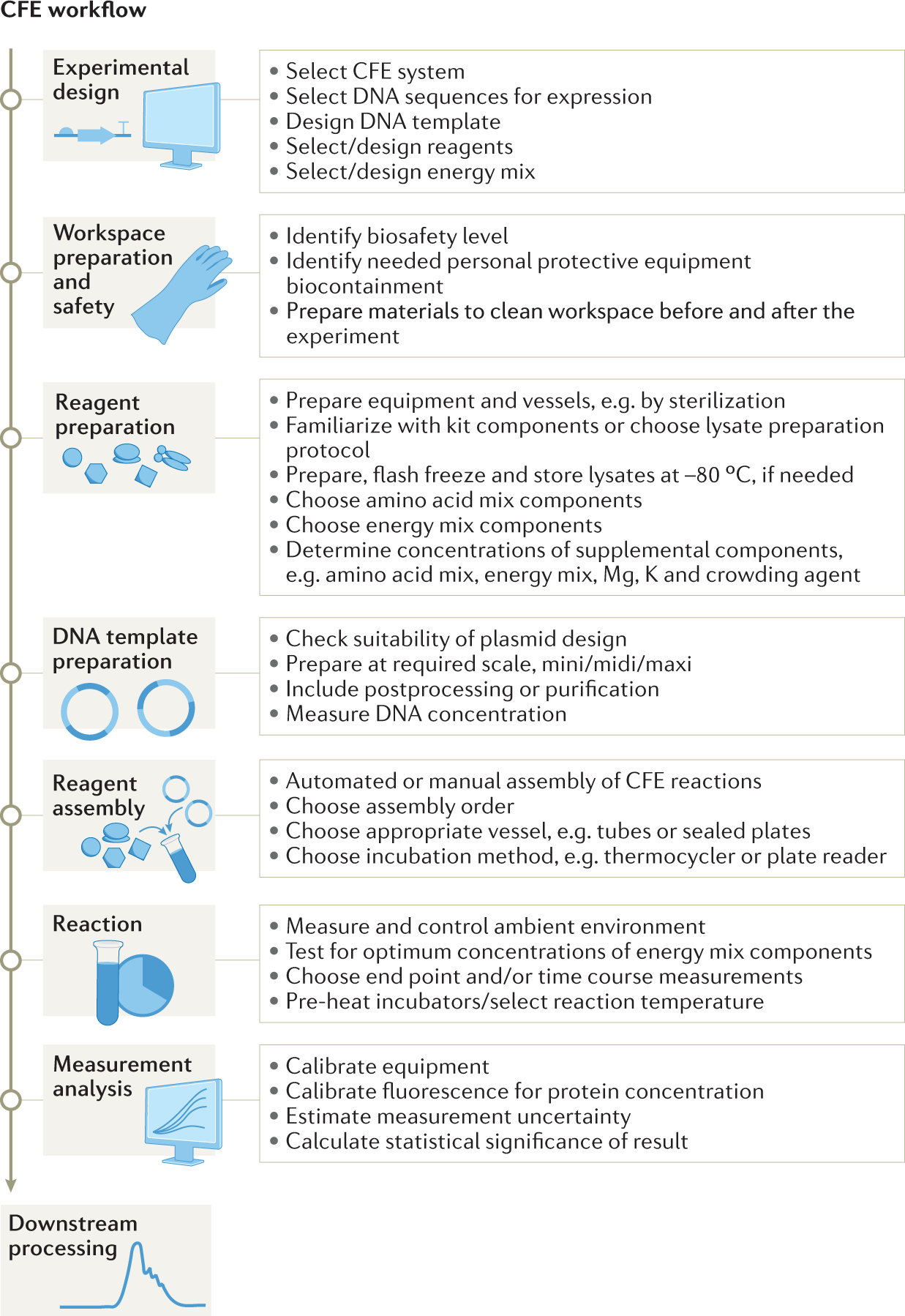
LC–MS-based quantification of intact proteins: perspective for clinical and bioanalytical applications | Bioanalysis

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero
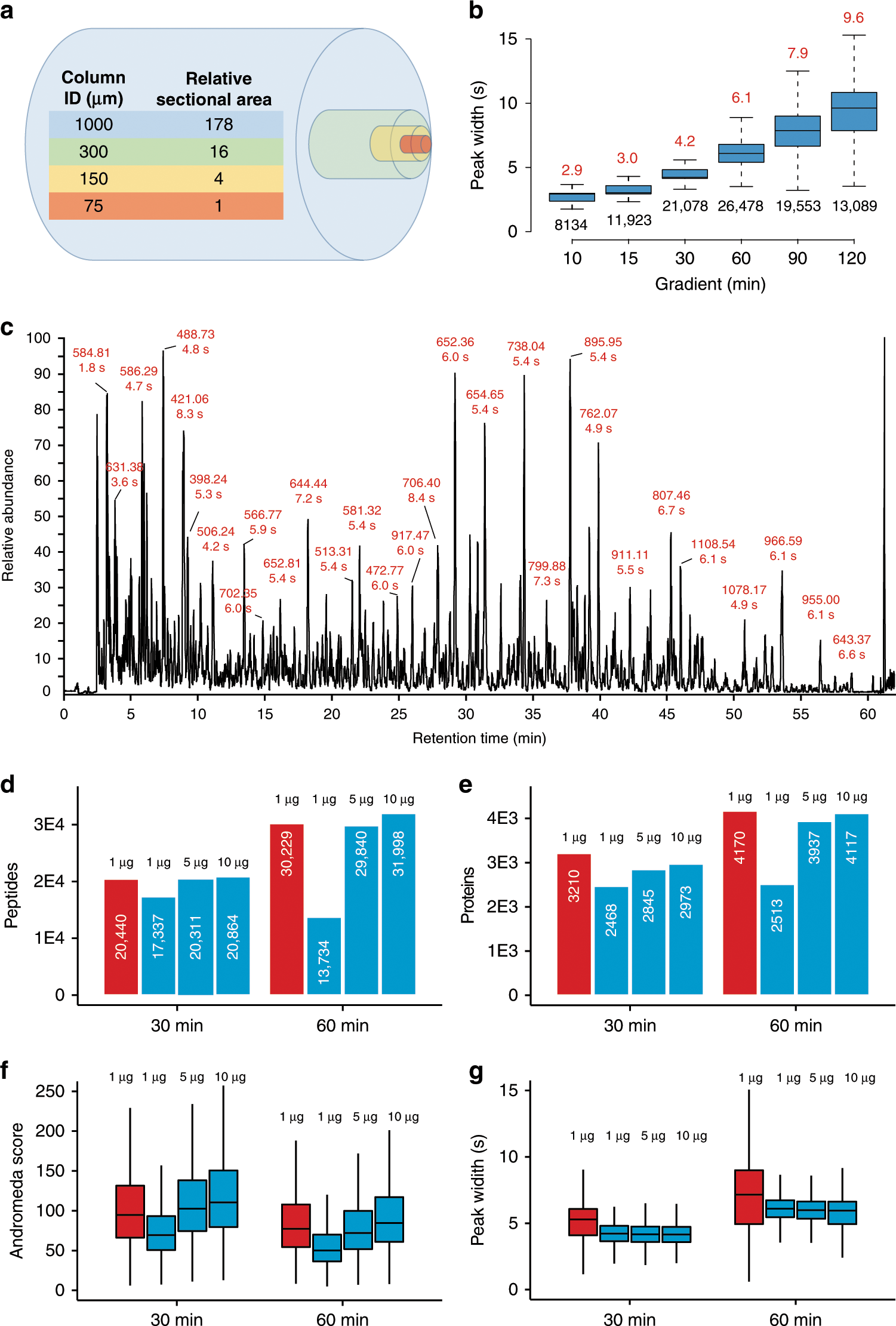
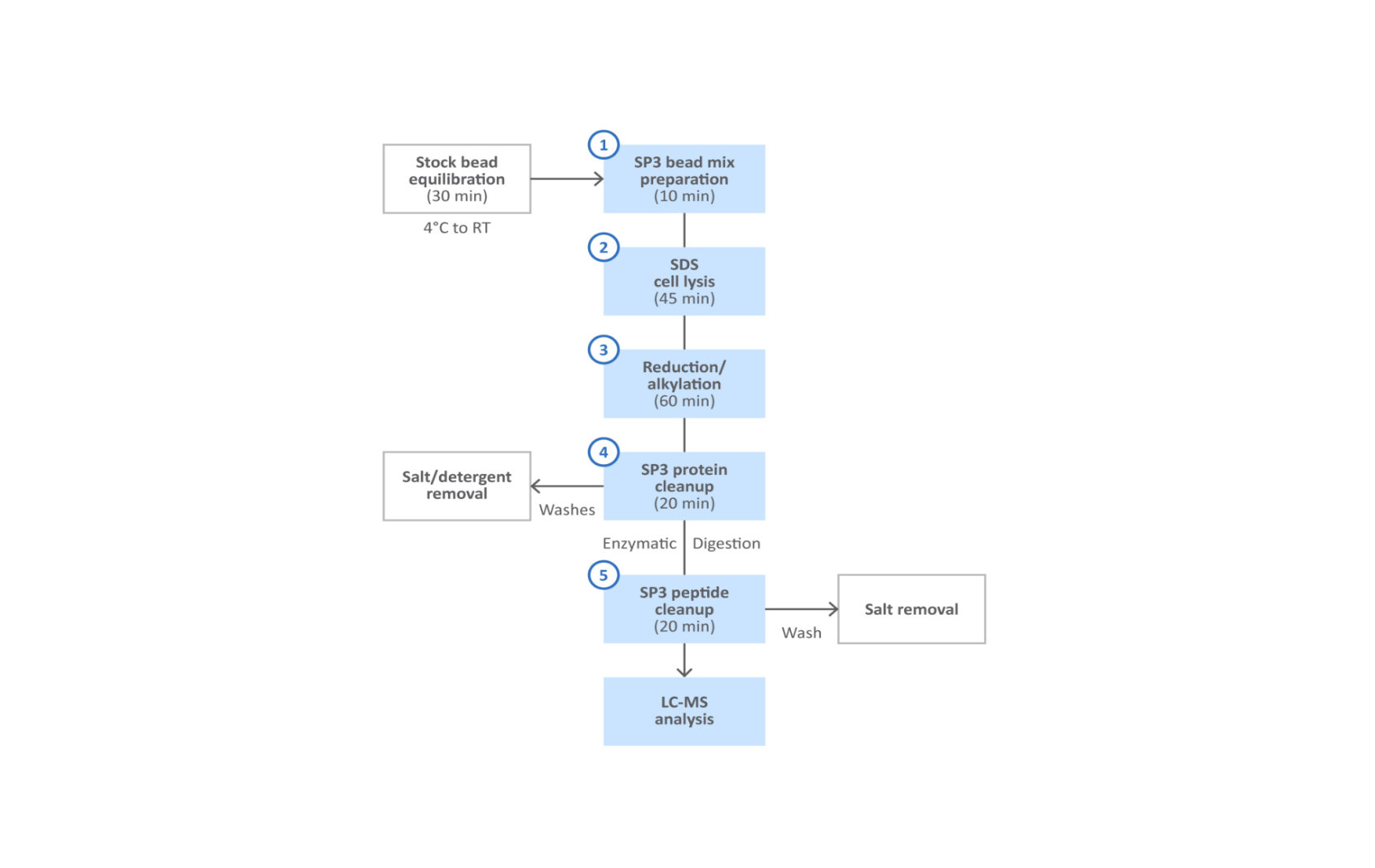
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications

Evaluation of pre-processing methods for tear fluid proteomics using proximity extension assays | Scientific Reports

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

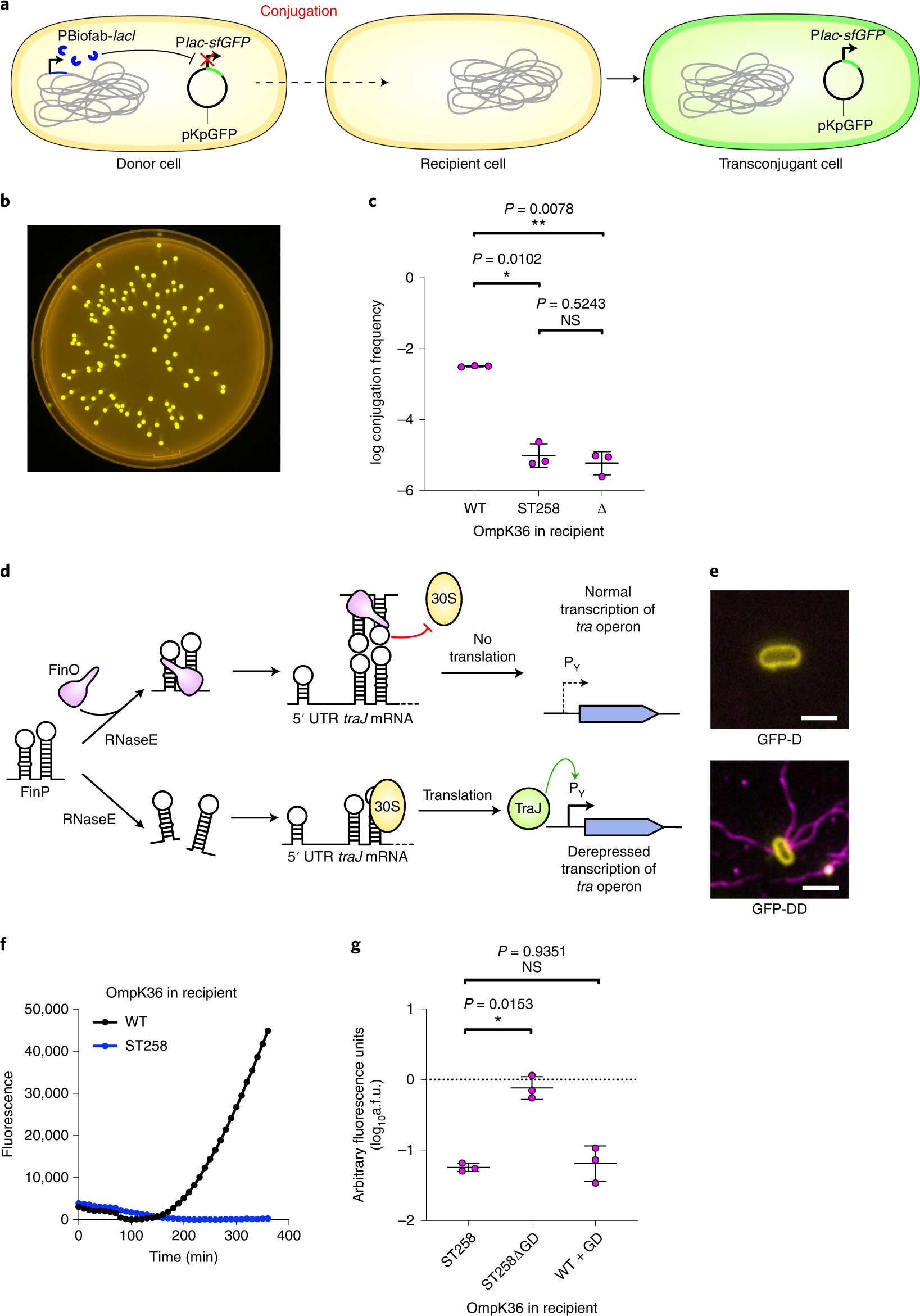
Quantitative Profiling of WNT-3A Binding to All Human Frizzled Paralogues in HEK293 Cells by NanoBiT/BRET Assessments | ACS Pharmacology & Translational Science

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

Single-Shot 10K Proteome Approach: Over 10,000 Protein Identifications by Data-Independent Acquisition-Based Single-Shot Proteomics with Ion Mobility Spectrometry | Journal of Proteome Research
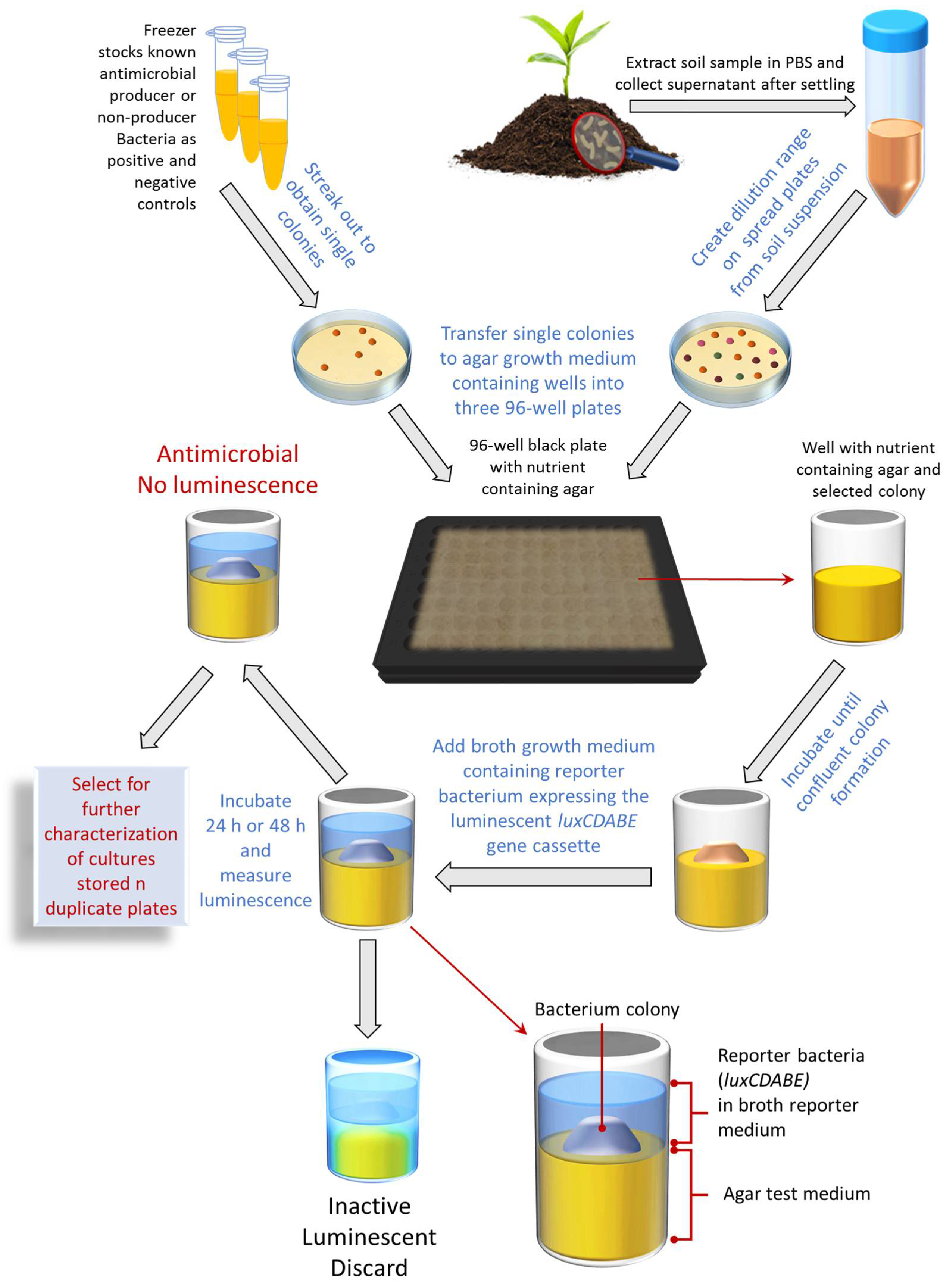
Microorganisms | Free Full-Text | Direct Detection of Antibacterial-Producing Soil Isolates Utilizing a Novel High-Throughput Screening Assay
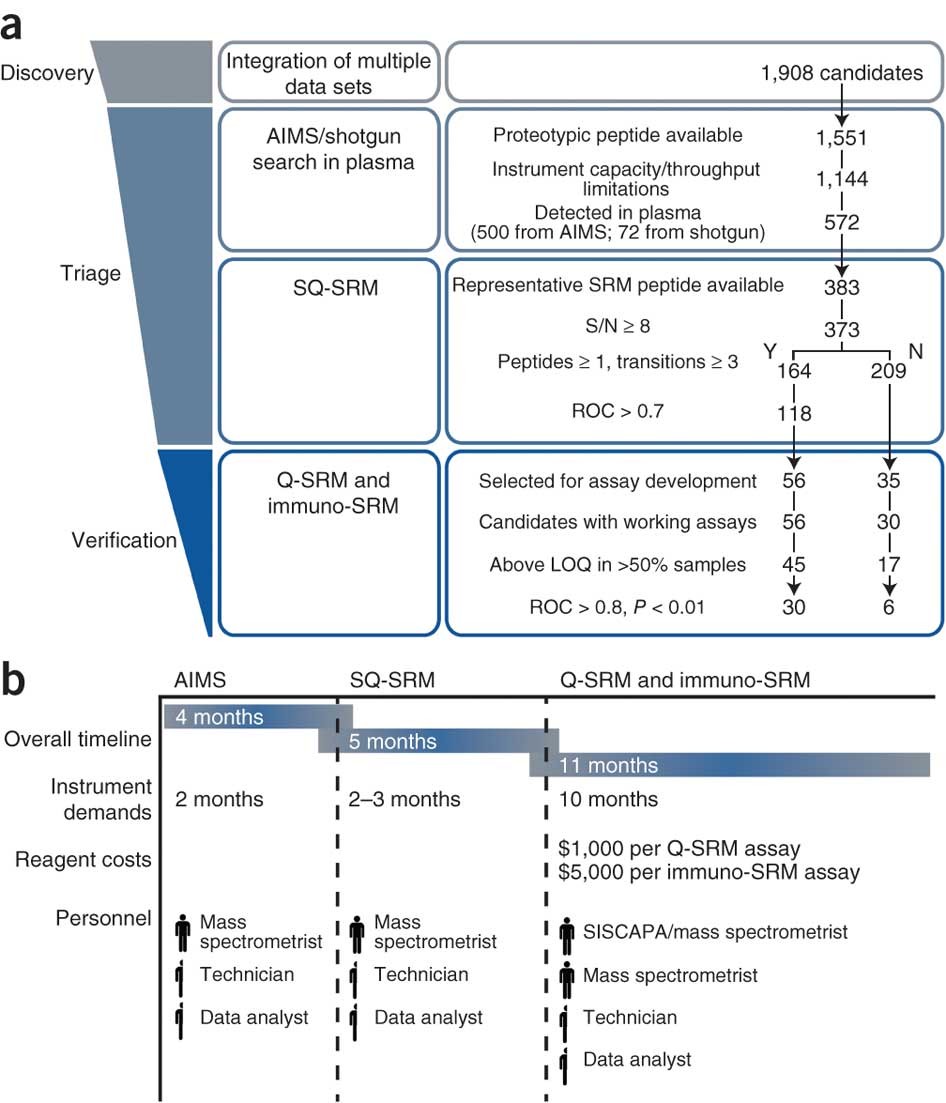
A targeted proteomics–based pipeline for verification of biomarkers in plasma | Nature Biotechnology

TR0068-Protein-assay-compatibility.pdf - TECH TIP #68 Protein assay compatibility table TR0068.5 Introduction No single protein assay method is | Course Hero

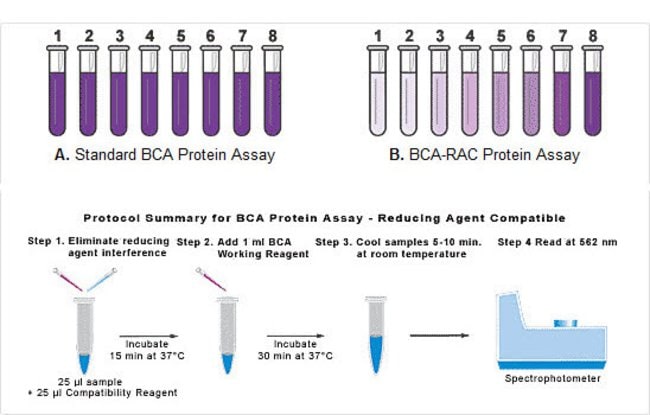
Fillable Online Protein assay compatibility table - Thermo Fisher Scientific Fax Email Print - pdfFiller

Reducing water uptake into BY‐2 cells by systematically optimizing the cultivation parameters increases product yields achieved by transient expression in plant cell packs - Opdensteinen - 2022 - Biotechnology Journal - Wiley Online Library