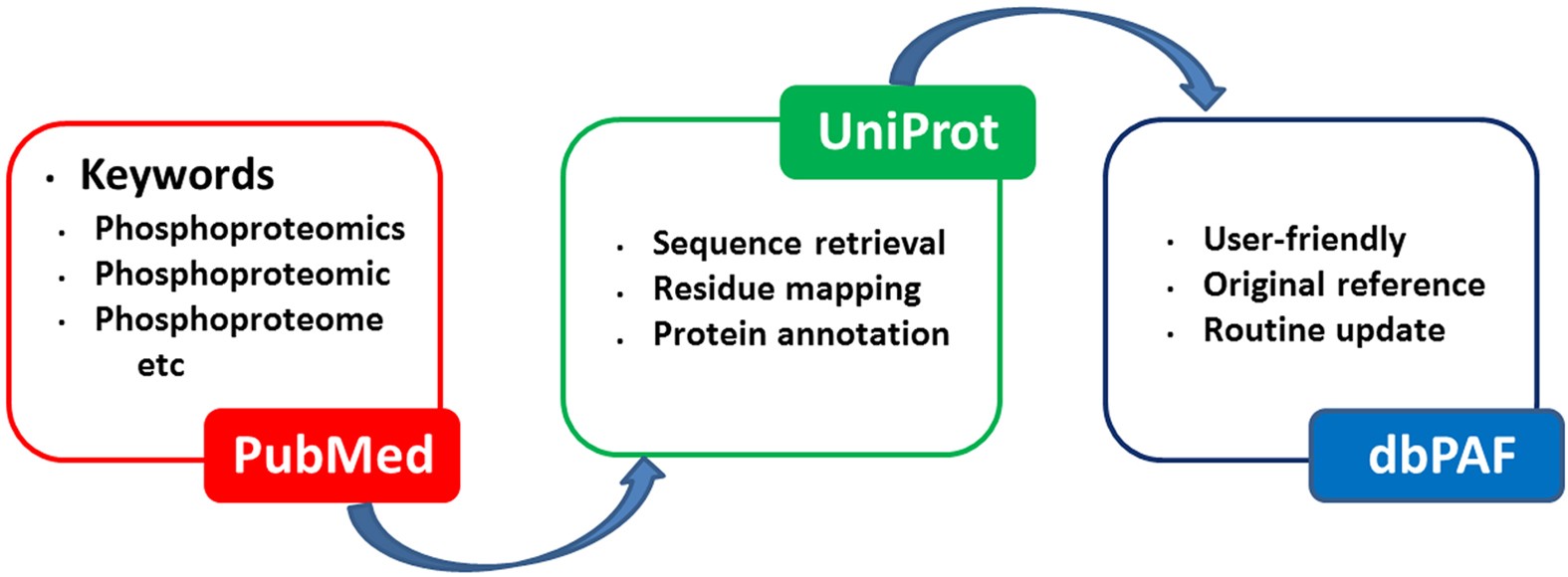
PhosphoPredict: A bioinformatics tool for prediction of human kinase-specific phosphorylation substrates and sites by integrating heterogeneous feature selection | Scientific Reports

Comparative Analysis Reveals Conserved Protein Phosphorylation Networks Implicated in Multiple Diseases | Science Signaling

Mapping and analysis of phosphorylation sites: a quick guide for cell biologists | Molecular Biology of the Cell

The switches.ELM Resource: A Compendium of Conditional Regulatory Interaction Interfaces | Science Signaling
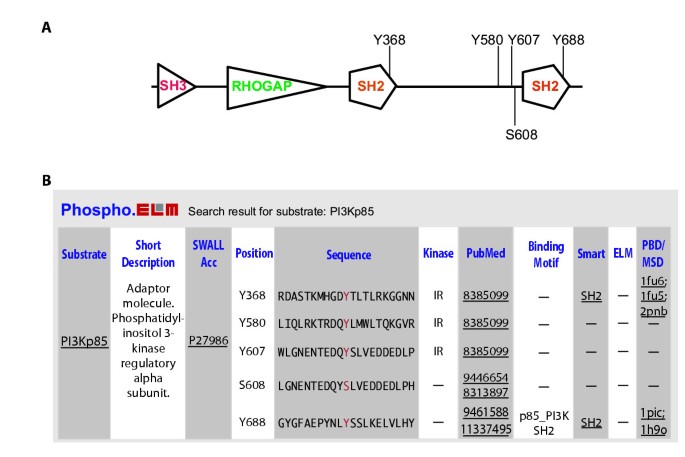
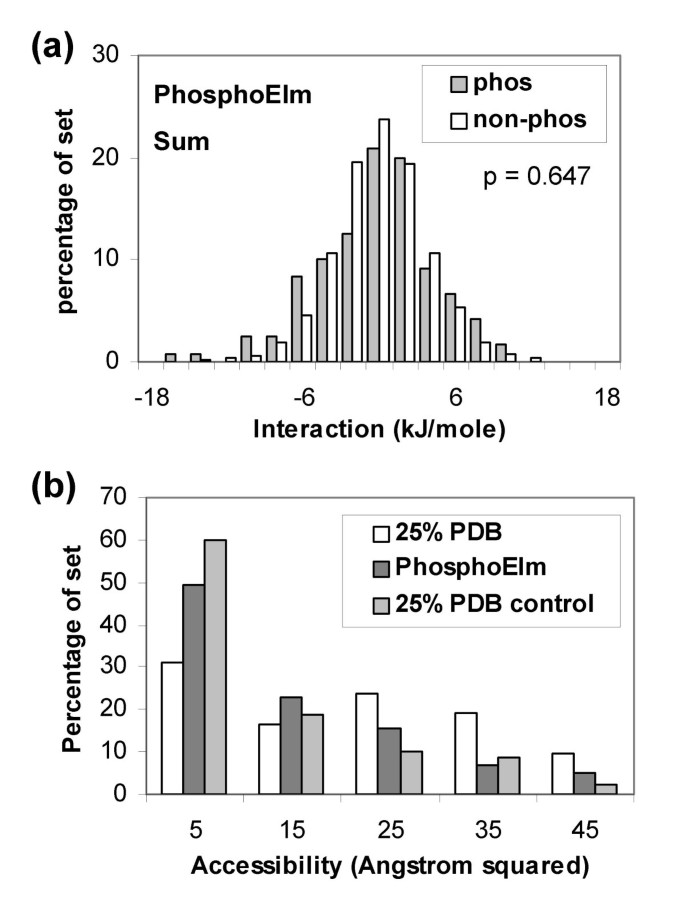
Phospho.ELM: A database of experimentally verified phosphorylation sites in eukaryotic proteins | BMC Bioinformatics | Full Text

Presentation of Phospho.ELM with the example of BLAST Search feature. a... | Download Scientific Diagram

Exploring protein phosphorylation by combining computational approaches and biochemical methods - ScienceDirect

IJMS | Free Full-Text | GasPhos: Protein Phosphorylation Site Prediction Using a New Feature Selection Approach with a GA-Aided Ant Colony System

Profiling the Human Phosphoproteome to Estimate the True Extent of Protein Phosphorylation | Journal of Proteome Research

Musite, a Tool for Global Prediction of General and Kinase-specific Phosphorylation Sites - ScienceDirect