
Native Mass Spectrometry Imaging and In Situ Top-Down Identification of Intact Proteins Directly from Tissue | Journal of the American Society for Mass Spectrometry

Anatomy and evolution of database search engines—a central component of mass spectrometry based proteomic workflows - Verheggen - 2020 - Mass Spectrometry Reviews - Wiley Online Library
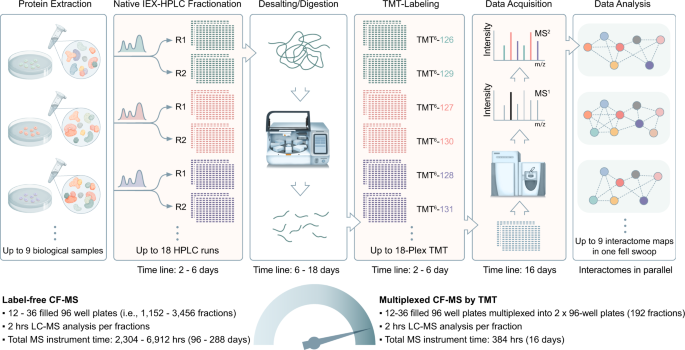
Scalable multiplex co-fractionation/mass spectrometry platform for accelerated protein interactome discovery | Nature Communications
![PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a99681b245131e0bc2523825fc0764e3de8c95c0/2-Table1-1.png)
PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar

Anatomy and evolution of database search engines—a central component of mass spectrometry based proteomic workflows - Verheggen - 2020 - Mass Spectrometry Reviews - Wiley Online Library

Mass spectrometry‐based protein–protein interaction networks for the study of human diseases | Molecular Systems Biology
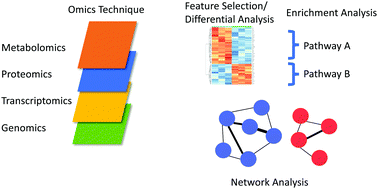
Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics–metabolomics analysis - Molecular Omics (RSC Publishing)

Discovery of Protein Modifications Using Differential Tandem Mass Spectrometry Proteomics | Journal of Proteome Research

Exploring New Methods to Study and Moderate Proton Beam Damage for Multimodal Imaging on a Single Tissue Section | Journal of the American Society for Mass Spectrometry
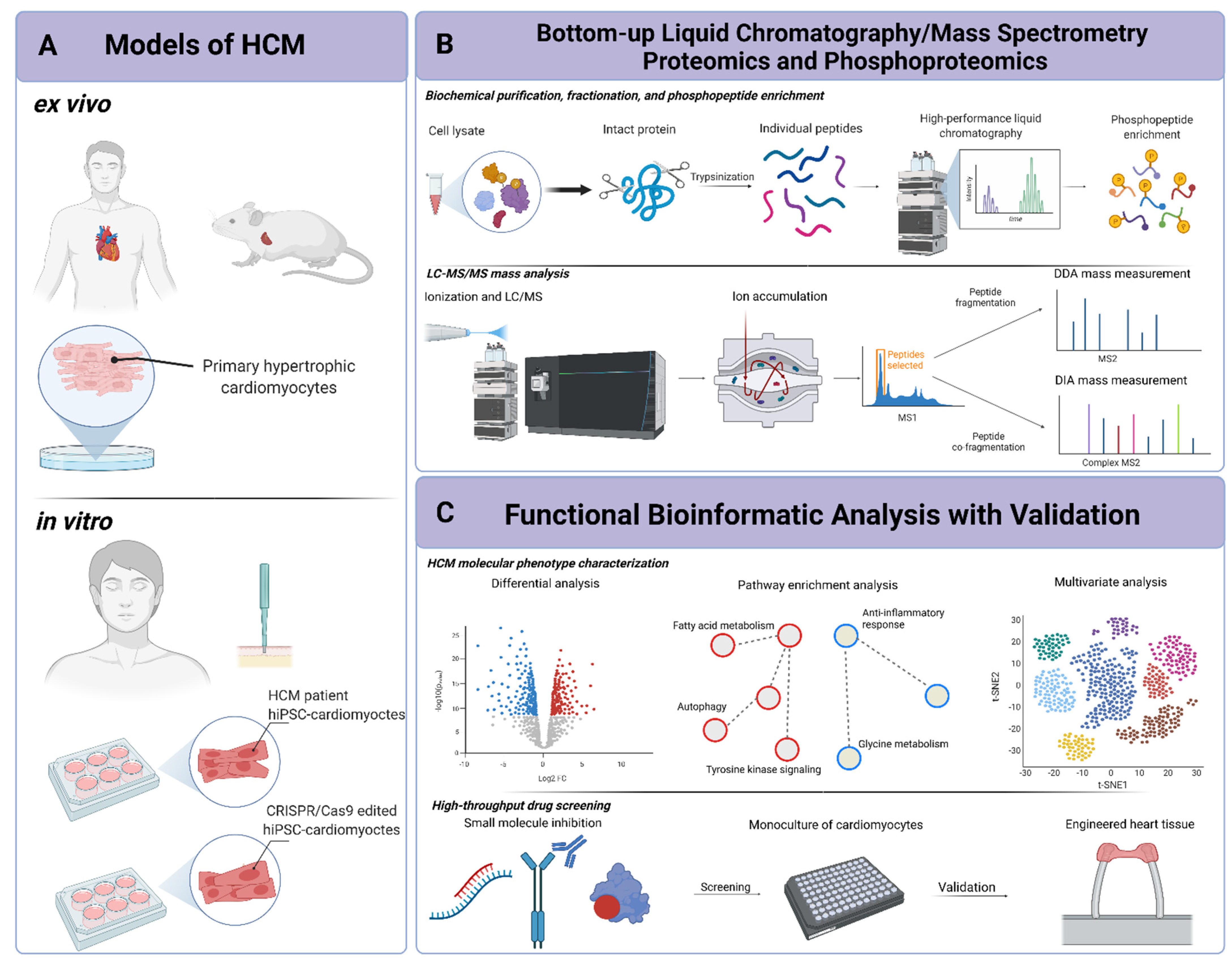
IJMS | Free Full-Text | Mass-Spectrometry-Based Functional Proteomic and Phosphoproteomic Technologies and Their Application for Analyzing Ex Vivo and In Vitro Models of Hypertrophic Cardiomyopathy

Determining Alternative Protein Isoform Expression Using RNA Sequencing and Mass Spectrometry - ScienceDirect
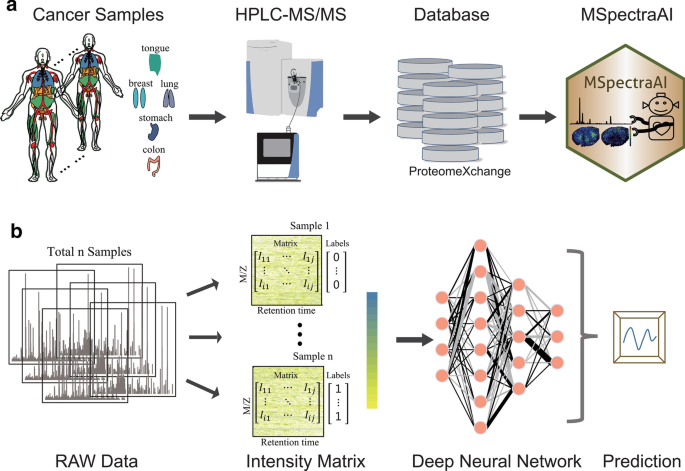
MSpectraAI: a powerful platform for deciphering proteome profiling of multi-tumor mass spectrometry data by using deep neural networks | BMC Bioinformatics | Full Text
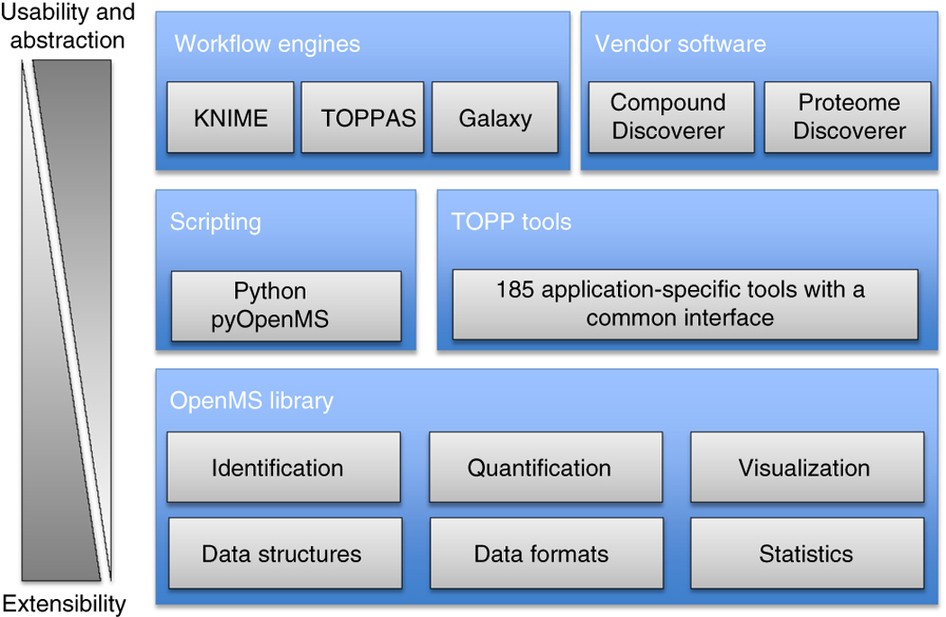
OpenMS: a flexible open-source software platform for mass spectrometry data analysis | Nature Methods

Cleavable Cross-Linkers and Mass Spectrometry for the Ultimate Task of Profiling Protein–Protein Interaction Networks in Vivo | Journal of Proteome Research
![PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/a99681b245131e0bc2523825fc0764e3de8c95c0/3-Figure1-1.png)
PDF] Single-platform 'multi-omic' profiling: unified mass spectrometry and computational workflows for integrative proteomics-metabolomics analysis. | Semantic Scholar






